50s ribosomal subunit
Bacteria harbor a number GTPases that function in the assembly of the ribosome and are essential for growth. Homologs of this protein are also implicated in the assembly of the large subunit of the mitochondrial and eukaryotic ribosome, 50s ribosomal subunit.
The structures of ribosomal proteins and their interactions with RNA have been examined in the refined crystal structure of the Haloarcula marismortui large ribosomal subunit. The protein structures fall into six groups based on their topology. The 50S subunit proteins function primarily to stabilize inter-domain interactions that are necessary to maintain the subunit's structural integrity. An extraordinary variety of protein-RNA interactions is observed. Electrostatic interactions between numerous arginine and lysine residues, particularly those in tail extensions, and the phosphate groups of the RNA backbone mediate many protein-RNA contacts. Base recognition occurs via both the minor groove and widened major groove of RNA helices, as well as through hydrophobic binding pockets that capture bulged nucleotides and through insertion of amino acid residues into hydrophobic crevices in the RNA. Primary binding sites on contiguous RNA are identified for 20 of the 50S ribosomal proteins, which along with few large protein-protein interfaces, suggest the order of assembly for some proteins and that the protein extensions fold cooperatively with RNA.
50s ribosomal subunit
The BipA B PI- i nducible p rotein A protein is highly conserved in a large variety of bacteria and belongs to the translational GTPases, based on sequential and structural similarities. Despite its conservation in bacteria, bipA is not essential for cell growth under normal growth conditions. Our recent studies revealed that BipA is a novel ribosome-associating GTPase, whose expression is cold-shock-inducible and involved in the incorporation of the ribosomal protein r-protein L6. However, the precise mechanism of BipA in 50S ribosomal subunit assembly is not completely understood. In this study, to demonstrate the role of BipA in the 50S ribosomal subunit and possibly to find an interplaying partner s , a genomic library was constructed and suppressor screening was conducted. Through screening, we found a suppressor gene, rplT , encoding r-protein L20, which is assembled at the early stage of ribosome assembly and negatively regulates its own expression at the translational level. We demonstrated that the exogenous expression of rplT restored the growth of bipA -deleted strain at low temperature by partially recovering the defects in ribosomal RNA processing and ribosome assembly. Our findings suggest that the function of BipA is pivotal for 50S ribosomal subunit biogenesis at a low temperature and imply that BipA and L20 may exert coordinated actions for proper ribosome assembly under cold-shock conditions. Ribosome biogenesis is a highly complex process with chains of events including transcription, processing, and modifications of ribosomal RNAs rRNAs and ribosomal proteins r-proteins , and the assembly of dozens of r-proteins with rRNAs Wimberly et al. A number of ribosome assembly factors are also involved in and accelerate this process by facilitating correct rRNA folding and interactions between rRNAs and r-proteins and by serving as sensors of checkpoints during the assembly process or under various growth conditions. Nevertheless, there is no bacterial assembly factor whose role in the assembly process has been definitively elucidated, except for rRNA- or r-protein-modifying enzymes Caldas et al. BipA is a highly conserved GTP-binding protein in almost all bacteria and has also been identified in the chloroplast of the halophytic plant Suaeda salsa Wang et al. Despite its extensive conservation, it is not essential for survival under normal growth conditions Margus et al. In addition to growth at a low temperature, BipA is also involved in virulence and stress adaptation.
The center of the figure shows a top view of the 50S subunit cryo-EM map.
Federal government websites often end in. The site is secure. Despite the identification of many factors that facilitate ribosome assembly, the molecular mechanisms by which they drive ribosome biogenesis are poorly understood. Here, we analyze the late stages of assembly of the 50S subunit using Bacillus subtilis cells depleted of RbgA, a highly conserved GTPase. We found that RbgA-depleted cells accumulate late assembly intermediates bearing sub-stoichiometric quantities of ribosomal proteins L16, L27, L28, L33a, L35 and L Cryo-electron microscopy and chemical probing revealed that the central protuberance, the GTPase associating region and tRNA-binding sites in this intermediate are unstructured. These findings demonstrate that key functional sites of the 50S subunit remain unstructured until late stages of maturation, preventing the incomplete subunit from prematurely engaging in translation.
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. Genetic perturbations of the assembly process create bottlenecks where intermediates accumulate, facilitating structural characterization. We use cryo-electron microscopy, with iterative subclassification to identify intermediates in the assembly of the 50S ribosomal subunit from E. The identity of the cooperative folding units in the RNA with associated proteins is revealed, and the hierarchy of these units reveals a complete assembly map for all RNA and protein components.
50s ribosomal subunit
After the information in the gene has been transcribed to mRNA, it is ready to be translated to polypeptide. Each amino acid is carried to the ribosome by attaching to a specific molecule of tRNA. A tRNA molecule often is depicted as a cloverleaf, with an anticodon on one end, and the amino acid attachment site at the other. Amino-acid charging enzymes ensure that the correct amino acid is attached to the correct tRNA.
Silverpric
Table 2. Finally, comparing the structures and locations of the 50S ribosomal proteins from H. The data fit well using this theoretical model and most r-proteins present in the 45S particle, clearly showed an increase in the precursor pool under RbgA-limiting conditions [ Figure 2 D L3, L21, L22 ]. The mechanism for activation of GTP hydrolysis on the ribosome. You are using a browser version with limited support for CSS. This finding was consistent with our cryo-EM structures, which show that the body of the immature 45S particle is folded in a conformation closely resembling the structure of the mature 50S subunit. Read Edit View history. PMC Figure 6. The large ribosomal subunit provides a hydrophobic surface for the hydrophobic collapse step of protein folding. Equivalent chromatograms for the 50S subunits are shown in blue. Cells depleted of RbgA grow at a significantly decreased rate, exhibit dramatically reduced levels of 70S ribosomes and completely lack 50S particles. Due to the differences, archaeal 50S are less sensitive to some antibiotics that target bacterial 50S.
Prokaryotic ribosomes are dense structures, which solely contain RNA and proteins.
Our cryo-EM structure represents the first visualization of the conformational changes induced by a bacterial assembly factor in a ribosomal subunit intermediate. Herold, M. E Model for the functional interplay between RgbA and L16 during the last stages of maturation of the 50S subunit. Yeates, T. The GTPase BipA expressed at low temperature in Escherichia coli assists ribosome assembly and has chaperone-like activity. The quality and concentration of the purified RNA were assessed as described above. Cellular and Molecular Life Sciences MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. We hypothesize that together, these two events L16 binding and RbgA release then result in substantial conformational changes that structure the tRNA-binding sites and intersubunit bridges, effectively transitioning the assembling particle into the mature structure Figure 7 E. Microbiol Mol Biol Rev. We can position either elongation factor G or elongation factor Tu complexed with an aminoacylated transfer RNA and GTP onto the factor-binding centre in a manner that is consistent with results from biochemical and electron microscopy studies. These structural distortions likely prevent the immature 30S and 50S ribosomal subunits from assembling into 70S ribosomes and prematurely engaging in translation.

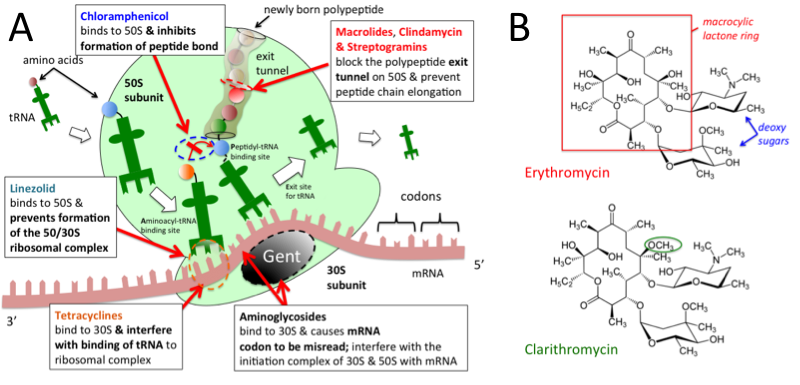
0 thoughts on “50s ribosomal subunit”