Clin var
Thank you for visiting nature.
Genome Medicine volume 15 , Article number: 51 Cite this article. Metrics details. Curated databases of genetic variants assist clinicians and researchers in interpreting genetic variation. Yet, these databases contain some misclassified variants. It is unclear whether variant misclassification is abating as these databases rapidly grow and implement new guidelines. Using archives of ClinVar and HGMD, we investigated how variant misclassification has changed over 6 years, across different ancestry groups.
Clin var
Interpretations of the clinical significance of variants are submitted by clinical testing laboratories, research laboratories, expert panels and other groups. ClinVar aggregates data by variant-disease pairs, and by variant or set of variants. Data aggregated by variant are accessible on the website, in an improved set of variant call format files and as a new comprehensive XML report. ClinVar recently started accepting submissions that are focused primarily on providing phenotypic information for individuals who have had genetic testing. ClinVar continues to make improvements to its search and retrieval functions. Several new fields are now indexed for more precise searching, and filters allow the user to narrow down a large set of search results. ClinVar 1 , 2 is a freely available, public archive of human genetic variants and interpretations of their significance to disease. Assertions of the clinical significance of a variant or set of variants are submitted to ClinVar by clinical testing laboratories, research laboratories, locus-specific databases, expert panels and other groups. Submissions include a description of the variant s ; the condition for which the variant was interpreted; the interpretation of the clinical significance of the variant, with the option to provide mode of inheritance; and evidence for that interpretation. ClinVar aggregates submissions based both on the variant and the variant-condition pair, and calculates an aggregate interpretation to indicate whether there is consensus or disagreement among submitters for an interpretation. Review status is based on submission of the criteria used by the submitter to classify variants, consensus across submitters in the interpretation of the variant and whether an expert panel or practice guideline-providing group has interpreted the variant. While most records in ClinVar report germline observations, about variants include somatic observations.
In addition to searching ClinVar with any clin var term, users can also perform advanced, focused searches by defining the field in which to look for the query term. Cite this article Sharo, A. To create Fig.
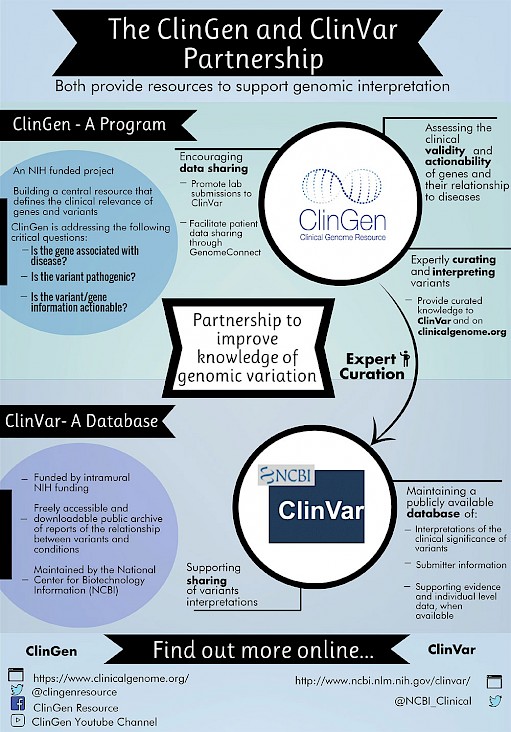
ClinVar and ClinGen, two NIH-based efforts, have formed a critical partnership to improve our knowledge of clinically relevant genomic variation. This partnership includes significant efforts in data sharing, data archiving, and collaborative curation to characterize and disseminate the clinical relevance of genomic variation. Share genomic and phenotypic data between clinicians, researchers, and patients through centralized and federated databases for clinical and research use. Develop and implement standards to support clinical annotation and interpretation of genes and variants. Develop data standards, software infrastructure and computational approaches to enable curation at scale and facilitate integration into healthcare delivery. When communicating the extremely close working relationship between ClinGen and ClinVar to researchers, clinicians, and the broader public, it is important to be clear and consistent. The following points should be highlighted:.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. Go to the search box in the gray area at the top of the page. Just type your search term and click on the Search button to the right of the search box.
Clin var
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. ClinVar is a freely accessible, public archive of reports of human variations classified for diseases and drug responses, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed conditions, and the history of those assertions.
The treadmill factory
Any term used to report clinical significance or phenotype that differs from standard is verified with the submitter. Wright et al. Brandi L Kattman. Since the P submission, GeneDx used variant classification criteria to classify this variant as VUS, citing the relatively high variant frequency as evidence for benignity. Researchers communicate variant classifications through published articles and submissions to variant databases. Skip to main content. Variant databases are under continuous development and growth [ 22 , 23 ]. We removed 5 variants that met the BA1 criteria in , 4 variants in , and 7 variants in Additional file 5 : Table S3. By December , 8 of these variants were reclassified to DM? The origin recognition complex requires chromatin tethering by a hypervariable intrinsically disordered region that is functionally conserved from sponge to man. We later discuss applying the BA1 guidelines. While many screened IEMs are debilitating or fatal in childhood unless treated, there are notable exceptions. However, this bias affecting genomes of African ancestry was no longer significant once common variants were removed in accordance with recent variant classification guidelines. While most records in ClinVar report germline observations, about variants include somatic observations.
.
Key ClinVar facts: ClinVar is fully public and freely available. These categories are defined in Stenson et al. Variant databases have taken different approaches to address misclassifications. Am J Hum Genetics. Blue lines indicate increasing pathogenicity or review stars, orange lines indicate increasing benignity or reduced confidence of pathogenicity, and gray lines indicate no change. Each ClinVar record represents the submitter, the variation and the phenotype, i. Genetic misdiagnoses and the potential for health disparities. Lee , George R. Evidence Evidence that supports an interpretation of the variation—phenotype relationship can be either highly structured or a free-text summary discussing how the evidence was evaluated. Evidence that supports an interpretation of the variation—phenotype relationship can be either highly structured or a free-text summary discussing how the evidence was evaluated. Data and source code for: ClinVar and HGMD genomic variant classification accuracy has improved over time, as measured by implied disease burden, Dryad, Dataset. This observation fits with clinical experience that mothers are more likely to bring a child to clinic and are therefore more likely to have similar clinical features observed and recorded. Variants within the same gene tended to be initially contributed by the same submitter.

In my opinion you are not right. I am assured. Let's discuss. Write to me in PM.
Many thanks for the information, now I will not commit such error.