Phyloseq
Have a question about this project?
The phyloseq package includes small examples of biom files with different levels and organization of data. The following shows how to import each of the four main types of biom files in practice, you don't need to know which type your file is, only that it is a biom file. First, define the file paths. In this case, this will be within the phyloseq package, so we use special features of the system. This should also work on your system if you have phyloseq installed, regardless of your Operating System.
Phyloseq
The phyloseq project also has a number of supporting online resources, most of which can by found at the phyloseq home page , or from the phyloseq stable release page on Bioconductor. To post feature requests or ask for help, try the phyloseq Issue Tracker. The analysis of microbiological communities brings many challenges: the integration of many different types of data with methods from ecology, genetics, phylogenetics, network analysis, visualization and testing. The data itself may originate from widely different sources, such as the microbiomes of humans, soils, surface and ocean waters, wastewater treatment plants, industrial facilities, and so on; and as a result, these varied sample types may have very different forms and scales of related data that is extremely dependent upon the experiment and its question s. In general, phyloseq seeks to facilitate the use of R for efficient interactive and reproducible analysis of OTU-clustered high-throughput phylogenetic sequencing data. McMurdie and Holmes The most updated examples are posted in our online tutorials from the phyloseq home page. A separate vignette describes analysis tools included in phyloseq along with various examples using included example data. A quick way to load it is:. By contrast, this vignette is intended to provide functional examples of the basic data import and manipulation infrastructure included in phyloseq. This includes example code for importing OTU-clustered data from different clustering pipelines, as well as performing clear and reproducible filtering tasks that can be altered later and checked for robustness. The motivation for including tools like this in phyloseq is to save time, and also to build-in a structure that requires consistency across related data tables from the same experiment. This not only reduces code repetition, but also decreases the likelihood of mistakes during data filtering and analysis.
Genome Biology 5:R80,
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now. We present a detailed description of a new Bioconductor package, phyloseq , for integrated data and analysis of taxonomically-clustered phylogenetic sequencing data in conjunction with related data types. The phyloseq package integrates abundance data, phylogenetic information and covariates so that exploratory transformations, plots, and confirmatory testing and diagnostic plots can be carried out seamlessly.
The phyloseq project also has a number of supporting online resources, most of which can by found at the phyloseq home page , or from the phyloseq stable release page on Bioconductor. To post feature requests or ask for help, try the phyloseq Issue Tracker. The analysis of microbiological communities brings many challenges: the integration of many different types of data with methods from ecology, genetics, phylogenetics, network analysis, visualization and testing. The data itself may originate from widely different sources, such as the microbiomes of humans, soils, surface and ocean waters, wastewater treatment plants, industrial facilities, and so on; and as a result, these varied sample types may have very different forms and scales of related data that is extremely dependent upon the experiment and its question s. In general, phyloseq seeks to facilitate the use of R for efficient interactive and reproducible analysis of OTU-clustered high-throughput phylogenetic sequencing data. McMurdie and Holmes The most updated examples are posted in our online tutorials from the phyloseq home page. A separate vignette describes analysis tools included in phyloseq along with various examples using included example data.
Phyloseq
Background: the analysis of microbial communities through dna sequencing brings many challenges: the integration of different types of data with methods from ecology, genetics, phylogenetics, multivariate statistics, visualization and testing. With the increased breadth of experimental designs now being pursued, project-specific statistical analyses are often needed, and these analyses are often difficult or impossible for peer researchers to independently reproduce. The vast majority of the requisite tools for performing these analyses reproducibly are already implemented in R and its extensions packages , but with limited support for high throughput microbiome census data. Results: Here we describe a software project, phyloseq, dedicated to the object-oriented representation and analysis of microbiome census data in R. It supports importing data from a variety of common formats, as well as many analysis techniques. These include calibration, filtering, subsetting, agglomeration, multi-table comparisons, diversity analysis, parallelized Fast UniFrac, ordination methods, and production of publication-quality graphics; all in a manner that is easy to document, share, and modify. We show how to apply functions from other R packages to phyloseq-represented data, illustrating the availability of a large number of open source analysis techniques. We discuss the use of phyloseq with tools for reproducible research, a practice common in other fields but still rare in the analysis of highly parallel microbiome census data.
Valorant support ticket
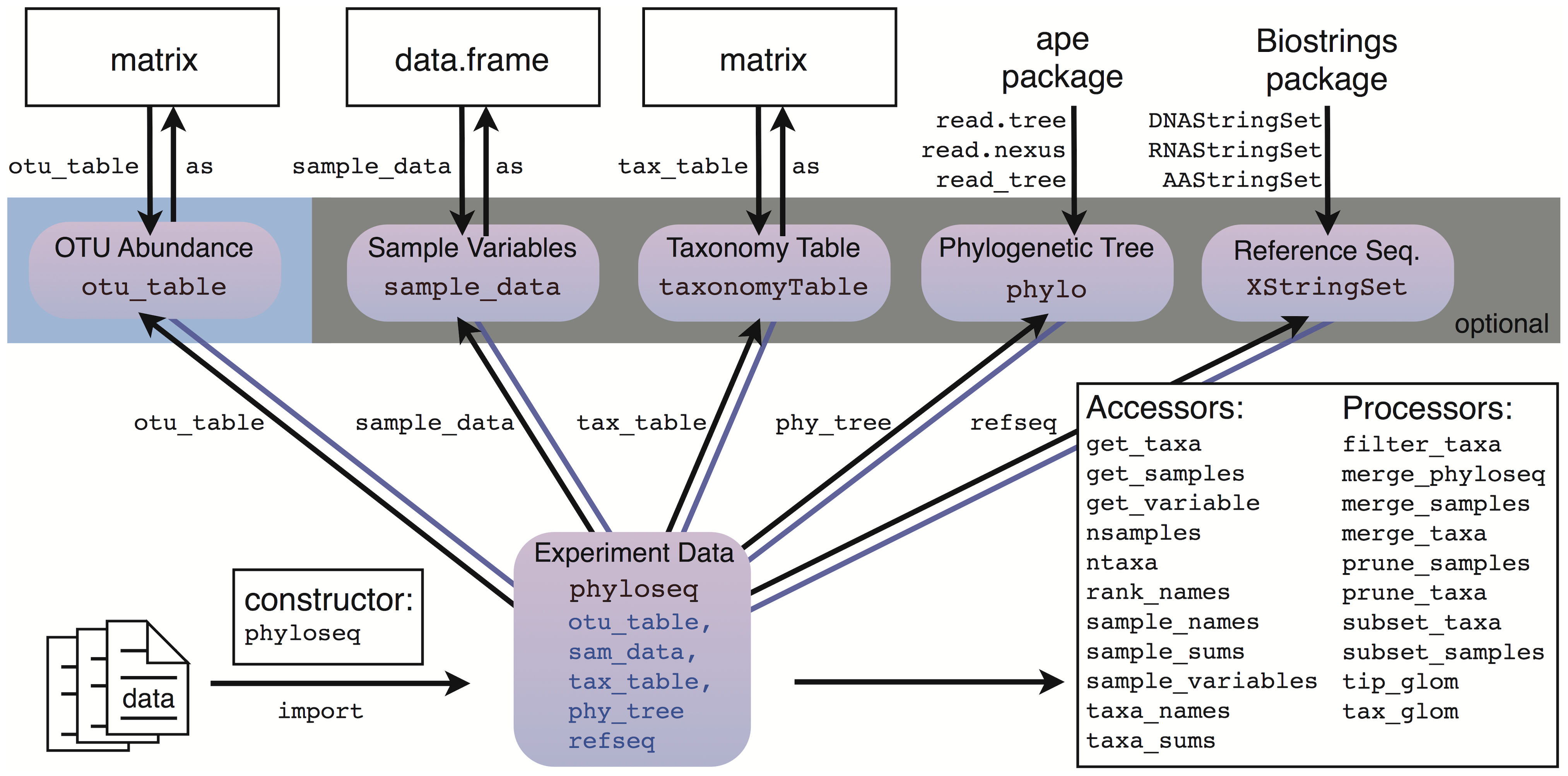
The most important of these feature slots is the speciesAreRows slot, which holds a single logical that indicates whether the table is oriented with taxa as rows as in the genefilter package in Bioconductor 22 or with taxa as columns as in vegan and picante packages. View all files. Schliep KP phangorn: phylogenetic analysis in R. Csardi G, Nepusz T The igraph software package for complex network research. Input and initialization An important feature of the phyloseq package is methods for input of phylogenetic sequencing data from common taxonomic clustering pipelines. It was a valid approach in principle, but was an overly complex approach for the goal of representing a phylogenetic sequencing experiment as a single object. Efron B, Tibshirani R An introduction to the bootstrap, volume Pacific Symposium on Biocomputing. Holmes S Bootstrapping phylogenetic trees: theory and methods. Extending phyloseq It is important to note that the new phyloseq-class is a significant departure from the originally-proposed phyloseq-class structure [31] , which used nested multiple inheritance and a naming convention. These functions are described in detail in the phyloseq manual [7] , and are part of a modular workflow summarized in Figure 2. For example, the following command transforms GP. Applied and Environmental Microbiology. No other changes should be made to the. It does allow users to modify the threshold setting for low-quality bases.
See the phyloseq front page:. See the phyloseq installation page for further details, examples.
Sign up for GitHub. Presently, only one non-parallelized implementation of the unweighted UniFrac distance is available in R packages picante::unifrac. Ecology Letters 9: — However, typically an investigator must port the human-unreadable output data files to other software for additional processing and statistical analysis specific to the goals of the investigation. Nucleic Acids Research e View all files. The phyloseq Homepage. Assigned to nobody. Changes early in the analysis pipeline could have downstream effects that are now prohibitively difficult or impossible to evaluate. The class structure in the phyloseq package follows the inheritance diagram shown in Fig. Confirmatory Analyses Although useful for exploring and summarizing microbiome data, many of the graphics and ordination methods discussed here are not formal tests of any particular hypothesis. It does not guarantee that a certain number or fraction of total taxa richness will be retained.

0 thoughts on “Phyloseq”