Race rapid amplification of cdna ends
The SMARTer protocol does not involve any of the adaptor ligation steps that other RACE kits incorporate, making the protocol shorter and significantly easier to execute. The Marathon cDNA Amplification Kit method employs a specially designed adaptor that significantly reduces background and permits both 5'- and 3'-RACE reactions Bertling, Beier, and Reichenberger ; Frohman to be performed using the same template. These nucleotides position the primer at the race rapid amplification of cdna ends of the poly A tail, eliminating the 3' heterogeneity inherent with conventional oligo dT priming. The UPM consists of two primers: a long, base primer and a short, base primer.
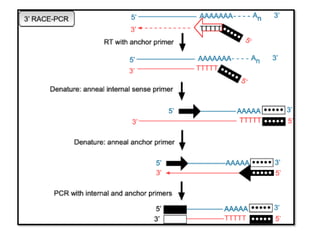
The amplified cDNA copies are then sequenced and, if long enough, should map to a unique genomic region. A more high-throughput alternative which is useful for identification of novel transcript structures, is to sequence the RACE-products by next generation sequencing technologies. In this process, an unknown end portion of a transcript is copied using a known sequence from the center of the transcript. The copied region is bounded by the known sequence, at either the 5' or 3' end. The primer binds to the mRNA, and the enzyme reverse transcriptase adds base pairs to the 3' end of the primer to generate a specific single-stranded cDNA product; this is the reverse complement of the mRNA. Following cDNA synthesis, the enzyme terminal deoxynucleotidyl transferase TdT is used to add a string of identical nucleotides , known as a homopolymeric tail, to the 3' end of the cDNA.
Race rapid amplification of cdna ends
Federal government websites often end in. The site is secure. The novel three-step protocol has been validated by mapping the transcriptional start sites of heterologously expressed yellow fever virus genomic RNAs from cultured mammalian cells. Determining the sequence of RNAs is a widely used routine in molecular biology, to which end mostly reverse transcription and consecutive polymerase chain reaction PCR 1 is employed. RACE procedures are commercially available in several customized versions in a kit format, mainly based on the strategies described in Refs. Finally, these PCR products are cloned in appropriate plasmid vectors for sequence analysis. For a detailed explanation, see text. As a key innovation, the RNA of interest is reverse-transcribed directly during the first step. Dots represent the consensus with the predicted sequence, and dashes represent nucleotide deletions. Dallmeier, unpublished to assess proper transcriptional start site selection [7,8] following plasmid DNA transfection into Vero-B African green monkey kidney cells as needed for productive viral RNA replication [9]. To that end, 0. Unreacted residual first-strand primers and cDNAs were removed by the addition of 1. The amplicon was visible as a single DNA species without multiple or unspecific side bands, much in contrast to what others have witnessed as a major drawback of inverted PCR on larger cDNAs several kilobases directly following circularization [10]. This treatment should render any noncapped RNA species sensitive to exonucleolytic degradation by Xrn-1 [11].
Wang C. The amplicon was visible as a single DNA species without multiple or unspecific side bands, much in contrast to what others have witnessed as a major drawback of inverted PCR on larger cDNAs several kilobases directly following circularization [10].
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. By using the oligonucleotide-containing random 9mer together with the GC-rich sequence for the suppression PCR technology at the first strand of cDNA synthesis, we have been able to amplify the cDNA from a very large transcript, such as the microtubule-actin crosslinking factor 1 MACF1 gene, which codes a transcript of 20 kb in size. Although the sequencing of the complete human genome revealed the presence of around 30,—40, genes Lander et al. Characterization of all splicing forms is essential to understand the function of each gene completely, since distinct splicing forms might have different functions Rahman et al.
The amplified cDNA copies are then sequenced and, if long enough, should map to a unique genomic region. A more high-throughput alternative which is useful for identification of novel transcript structures, is to sequence the RACE-products by next generation sequencing technologies. In this process, an unknown end portion of a transcript is copied using a known sequence from the center of the transcript. The copied region is bounded by the known sequence, at either the 5' or 3' end. The primer binds to the mRNA, and the enzyme reverse transcriptase adds base pairs to the 3' end of the primer to generate a specific single-stranded cDNA product; this is the reverse complement of the mRNA. Following cDNA synthesis, the enzyme terminal deoxynucleotidyl transferase TdT is used to add a string of identical nucleotides , known as a homopolymeric tail, to the 3' end of the cDNA. There are some other ways to add the 3'-terminal sequence for the first strand of the de novo cDNA synthesis which are much more efficient than homopolymeric tailing, but the sense of the method remains the same.
Race rapid amplification of cdna ends
Thank you for visiting nature. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript. The method consists of using PCR to amplify, from complex mixtures of cellular mRNA, the regions between the known parts of the sequence and non-specific tags appended to the ends of the cDNA. This is a preview of subscription content, access via your institution.
Why did linda leave blue bloods
J Mol Biol — This second set of primers further amplifies the cDNA of interest but not the non-specific product, increasing the specificity and yield of the reaction. C Suppression PCR technology. Typically, with novel mRNAs only a portion of its complete sequence is known. Next, use a microvolume spectrophotometer to measure the RNA concentration. To that end, 0. Following cDNA synthesis, the enzyme terminal deoxynucleotidyl transferase TdT is used to add a string of identical nucleotides , known as a homopolymeric tail, to the 3' end of the cDNA. To obtain the best experience, we recommend you use a more up to date browser or turn off compatibility mode in Internet Explorer. Certain trademarks may not be registered in all jurisdictions. Lane 1: 1. The sequence at the five-prime end can be amplified similarly. Natick, MA, pp —
Federal government websites often end in. The site is secure. We describe a novel method for the specific amplification of cDNA ends.
Before beginning the synthesis and amplification of the cDNA, use a primer design software to create a five-prime specific primer for the gene of interest, dSmad2 in this example. Moreover, by including suppression PCR technology Chen et al. This synthesized strand readily forms a panhandle-like structure after the denaturing step because this structure is more stable than the primer-template hybrid and therefore suppresses exponential amplification and reduces nonspecific products. Download references. Using these diluted products, prepare the second round amplification PCR mix. Incubate the reactions for 90 minutes at 42 degrees Celsius. The new kit is much more successful in amplifying strong, single bands across the sample set. E Sequence analysis of each PCR product using direct sequencing. References 1. In contrast, as shown in Fig. Please check your Internet connection and reload this page. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. See improved performance with a protocol designed to accommodate larger RNA input volumes and perform more efficiently on challenging targets.

I have not understood, what you mean?
I am sorry, it does not approach me. There are other variants?