Riboswitch
Federal government websites often end in. The site is secure, riboswitch. A riboswitch collection of bacterial riboswitch classes is being discovered that sense central metabolites, coenzymes, riboswitch, and signaling molecules. In this review, the mechanisms of riboswitch-mediated translation control are summarized to highlight both their diversity and potential ancient origins.
Federal government websites often end in. The site is secure. A critical feature of the hypothesized RNA world would have been the ability to control chemical processes in response to environmental cues. Riboswitches present themselves as viable candidates for a sophisticated mechanism of regulatory control in RNA-based life. In this review, we focus on recent insights into how these RNAs fold into complex architectures capable of both recognizing a specific small molecule compound and exerting regulatory control over downstream sequences, with an emphasis on transcriptional regulation.
Riboswitch
This page has been archived and is no longer updated. Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. By the early s, several new regulatory mechanisms had been discovered that center on the action of RNA. Arnaud et al. Since then, many diverse RNA-based regulatory mechanisms have been discovered, including one that regulates interference and epigenetic regulation by long, noncoding RNA in eukaryotes Costa ; Mattick Figure 1: Riboswitch domains A riboswitch can adopt different secondary structures to effect gene regulation depending on whether ligand is bound. This schematic is an example of a riboswitch that controls transcription. When metabolite is not bound -M , the expression platform incorporates the switching sequence into an antiterminator stem-loop AT and transcription proceeds through the coding region of the mRNA. One common form of riboregulation in bacteria is the use of ribonucleic acid sequences encoded within mRNA that directly affect the expression of genes encoded in the full transcript called cis -acting elements because they act on the same molecule they're coded in. Where are riboswitches located? There they regulate the occlusion of signals for transcription attenuation or translation initiation Figure 2. However, not all riboswitches are at the 5' UTR; scientists have discovered that, in some eukaryotic mRNA, the thiamine pyrophosphate TPP riboswitch regulates splicing at the 3' end Wachter et al. The function of riboswitches is tied to the ability of RNA to form a diversity of structures.
Mind Read.
Riboswitches are specific components of an mRNA molecule that regulates gene expression. The riboswitch is a part of an mRNA molecule that can bind and target small target molecules. An mRNA molecule may contain a riboswitch that directly regulates its own expression. The riboswitch displays the ability to regulate RNA by responding to concentrations of its target molecule. Hence, the existence of RNA molecules provide evidence to the RNA world hypothesis that RNA molecules were the original molecules, and that proteins developed later in evolution. Riboswitches are found in bacteria, plants, and certain types of fungi.
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now. A critical feature of the hypothesized RNA world would have been the ability to control chemical processes in response to environmental cues.
Riboswitch
This page has been archived and is no longer updated. Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. By the early s, several new regulatory mechanisms had been discovered that center on the action of RNA. Arnaud et al. Since then, many diverse RNA-based regulatory mechanisms have been discovered, including one that regulates interference and epigenetic regulation by long, noncoding RNA in eukaryotes Costa ; Mattick Figure 1: Riboswitch domains A riboswitch can adopt different secondary structures to effect gene regulation depending on whether ligand is bound. This schematic is an example of a riboswitch that controls transcription. When metabolite is not bound -M , the expression platform incorporates the switching sequence into an antiterminator stem-loop AT and transcription proceeds through the coding region of the mRNA. One common form of riboregulation in bacteria is the use of ribonucleic acid sequences encoded within mRNA that directly affect the expression of genes encoded in the full transcript called cis -acting elements because they act on the same molecule they're coded in.
Hemmings.com
As a second mechanism, when a protein binds specifically to the aptamer region, it can induce changes in the mRNA structure, including the formation or removal of inhibitory mRNA structures that the ribosome must unfold prior to initiating translation. After the ribosome has bound to form a pre-initiation complex, there are significant structural re-arrangements, including unfolding an inhibitory structure within the N-terminal CDS region 40 , It has been proposed that these intermediates provide the RNA with a mechanism for preventing misfolding and enhancing the rate of the overall folding reaction in vivo. Pyrithiamine pyrophosphate has been shown to bind and activate the TPP riboswitch, causing the cell to cease the synthesis and import of TPP. It would be exceedingly wasteful for the riboswitch to regulate the translation of only the first gene by occluding its RBS on AdoCbl binding, but still allow the cell to synthesize the entire mRNA and synthesize all the other 24 proteins encoded by this multi-ORF message. A complex set of interacting helices and loop formations defines the overall three-dimensional fold of the purine riboswitch aptamer domain where ligands bind Batey et al. Rogers, J. Figure 3. The switching mechanism for riboswitches of the type found in N. Cell , — Trends Biochem Sci 41 : — Gene Ther 16 : — As these processes are often cotranscriptional, it is important to consider the rate at which RNA transcribes and folds, as well as the relative thermodynamic stability of competing secondary structures.
Federal government websites often end in. The site is secure.
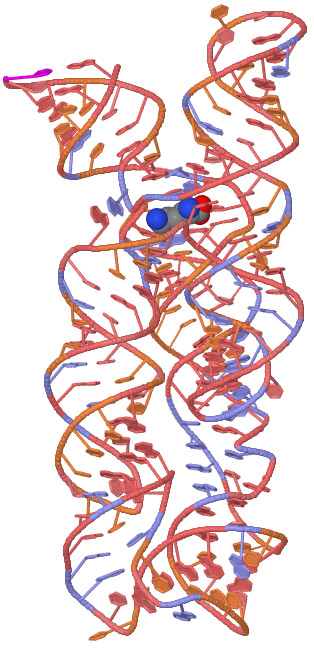
Purine Riboswitch Family: Global Structure. One of the most notable features of the ligand-bound aptamer domain is the way it nearly completely encapsulates the ligand, with the surrounding aqueous environment making the ligand almost entirely inaccessible Batey et al. Mayer G , Chap. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. This is a hallmark of a kinetically controlled process in which the aptamer domain has insufficient time to equilibrate with the cellular environment before the expression platform commits the RNA to an alternative folding route that may be largely irreversible. References and Recommended Reading Arnaud, M. The coupling of ligand binding to a conformational change is central to the regulatory mechanism of the riboswitch. Thavarajah, W. After completion of the aptamer domain, often a programmed pause site in the mRNA causes the polymerase to temporarily stall Figure 5, star. Riboswitches: The oldest mechanism for the regulation of gene expression? This tandem arrangement between a riboswitch aptamer and a self-splicing ribozyme exploits the typical activities of each RNA device to create a far more sophisticated gene-control apparatus.

In my opinion the theme is rather interesting. I suggest all to take part in discussion more actively.