Scispacy
A beginner's guide to using Named-Entity Recognition for data extraction from biomedical literature, scispacy. This code walks you through the installation and usage of scispaCy for natural language processing. For our example, we use scispacy from CORD, a large collection of scispacy about the Covid pandemic.
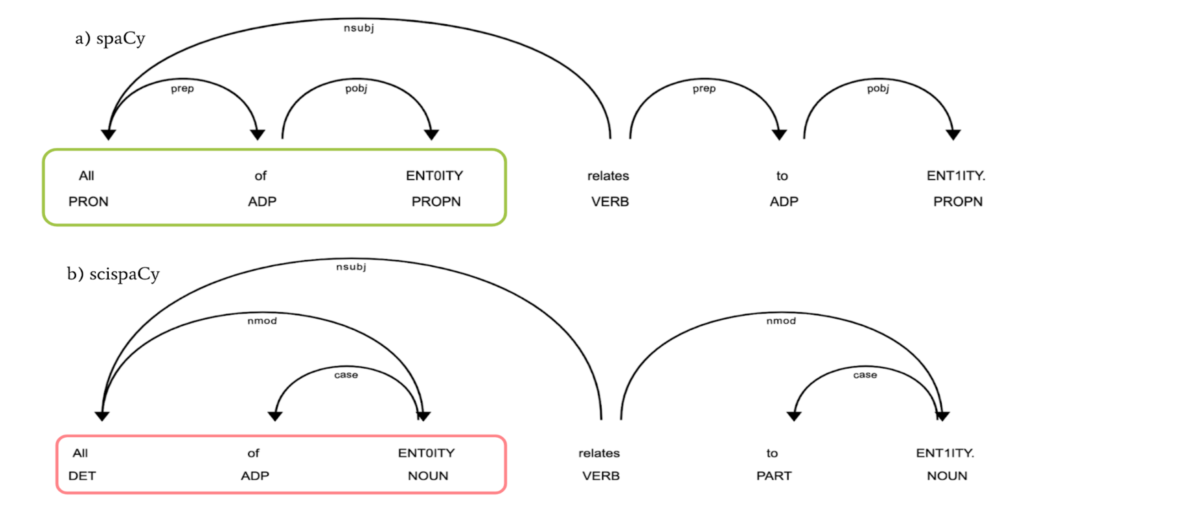
This repository contains custom pipes and models related to using spaCy for scientific documents. In particular, there is a custom tokenizer that adds tokenization rules on top of spaCy's rule-based tokenizer, a POS tagger and syntactic parser trained on biomedical data and an entity span detection model. Separately, there are also NER models for more specific tasks. Just looking to test out the models on your data? Check out our demo Note: this demo is running an older version of scispaCy and may produce different results than the latest version.
Scispacy
Released: Feb 20, View statistics for this project via Libraries. Author: Allen Institute for Artificial Intelligence. Tags bioinformatics, nlp, spacy, SpaCy, biomedical. Mar 8, Sep 30, Apr 29, Sep 7, Mar 10, Feb 12, Oct 16, Jul 8,
Last commit date.
.
Full Changelog : v0. Note: The models e. This release of scispacy is compatible with Spacy 3. This will come in a later release. This component produces a doc level attribute on the spacy doc: doc. The tuples contain:. Thanks to Yoav Goldberg for this fix! Yoav wrote some scripts to convert between them, including normalising of some syntactic phenomena that were being treated inconsistently between the two corpora. This contains a smaller set of higher quality entities, which are used for indexing in Pubmed. It is comprised of several other drug vocabularies commonly used in pharmacy management and drug interaction, including First Databank, Micromedex, and the Gold Standard Drug Database.
Scispacy
Despite recent advances in natural language processing, many statistical models for processing text perform extremely poorly under domain shift. Processing biomedical and clinical text is a critically important application area of natural language processing, for which there are few robust, practical, publicly available models. We detail the performance of two packages of models released in scispaCy and demonstrate their robustness on several tasks and datasets.
Abc consultants
Supported by. Example text before NER:. Take a look below in the "Setting up a virtual environment" section if you need some help with this. The models are installed using their URLs, found here. Example Usage. Packages 0 No packages published. Custom properties. Available Models. Notifications Fork 13 Star To install the library, run:. Reload to refresh your session. Once that is done, we pick a specific text to extract from that file and pass it through one of the models. Activate the Conda environment. You signed out in another tab or window.
In its most basic form a spaCy application can be very short, but a lot of processing steps take place, and a lot more information is contained within the doc object. If your result is a shorter list of pipeline components then you are likely not using the most recent version of spaCy.
Uploaded Feb 20, py3. In particular, there is a custom tokenizer that adds tokenization rules on top of spaCy's rule-based tokenizer, a POS tagger and syntactic parser trained on biomedical data and an entity span detection model. Author: Allen Institute for Artificial Intelligence. Additionally, scispacy uses modern features of Python and as such is only available for Python 3. Newer version available 0. If you are upgrading scispacy , you will need to download the models again, to get the model versions compatible with the version of scispacy that you have. Separately, there are also NER models for more specific tasks. Releases No releases published. Apr 3, View all files.

I apologise, but, in my opinion, you commit an error.
I think, that you are mistaken. I suggest it to discuss. Write to me in PM, we will talk.