Clinvar
ClinVar aggregates information about genomic variation and its relationship to clinvar health, clinvar. ClinVar GitHub. We will release changes to the ClinVar XML files and our submission spreadsheet templates on January 29 ; clinvar changes will improve support for classifications of somatic variants in ClinVar. To help file submitters prepare for this change, clinvar, we are making the updated spreadsheet templates available for clinvar with a note explaining changes.
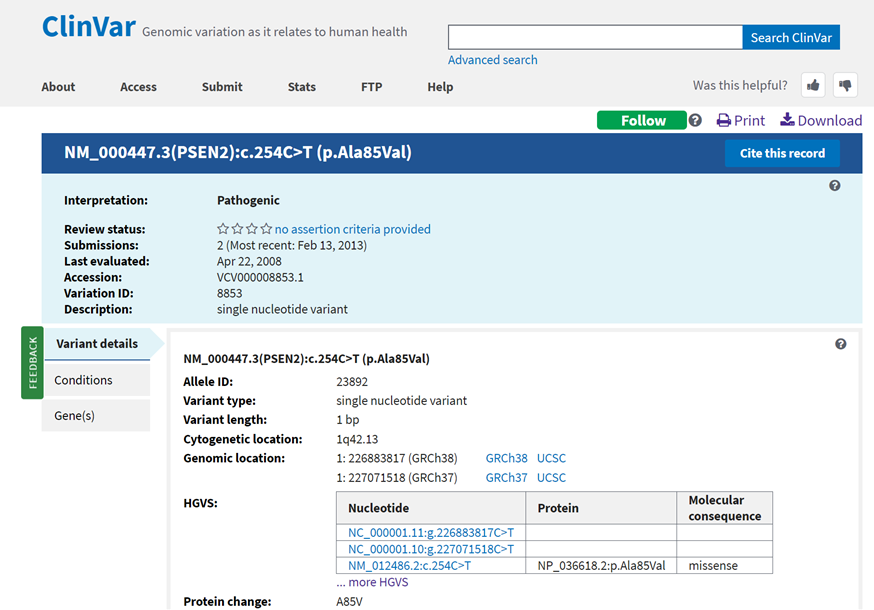
Federal government websites often end in. The site is secure. ClinVar accessions submissions reporting human variation, interpretations of the relationship of that variation to human health and the evidence supporting each interpretation. The database is tightly coupled with dbSNP and dbVar, which maintain information about the location of variation on human assemblies. Each ClinVar record represents the submitter, the variation and the phenotype, i. The submitter can update the submission at any time, in which case a new version is assigned.
Clinvar
The content on this website is based on ClinVar database version July 14, Simple ClinVar was developed to provide gene- and disease-wise summary statistic based on all available genetic variants from ClinVar. How many missense variants are associated to heart disease? What are the top 10 genes mutated in Alzheimer? Does CDKL5 have pathogenic mutations? If so, where? Simple ClinVar is able to answer these questions and more, in a matter of seconds. For detailed information about Simple ClinVar please refer to the original publication:. Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database. Clinical genetic testing has exponentially expanded in recent years, leading to an overwhelming amount of patient variants with high variability in pathogenicity and heterogeneous phenotypes.
The database query mode coupled with dynamic filtering allows the user to explore which are the most common clinvar and types of variants most commonly found in the whole database. As of Decemberclinvar, 10 of these 11 variants have been reclassified in ClinVar to a non-pathogenic category.
NOTE: ClinVar is intended for use primarily by physicians and other professionals concerned with genetic disorders, by genetics researchers, and by advanced students in science and medicine. While the ClinVar database is open to all academic users, users seeking information about a personal medical or genetic condition are urged to consult with a qualified physician for diagnosis and for answers to personal questions. These tracks show the genomic positions of variants in the ClinVar database. ClinVar is a free, public archive of reports of the relationships among human variations and phenotypes, with supporting evidence. Because the ClinVar type no longer captures this information, any variation equal to or larger than 50 bp is now considered a CNV. The ClinVar Interpretations track displays the genomic positions of individual variant submissions and interpretations of the clinical significance and their relationship to disease in the ClinVar database. Variants may be right-aligned or may contain additional context, e.
The content on this website is based on ClinVar database version July 14, Simple ClinVar was developed to provide gene- and disease-wise summary statistic based on all available genetic variants from ClinVar. How many missense variants are associated to heart disease? What are the top 10 genes mutated in Alzheimer? Does CDKL5 have pathogenic mutations? If so, where?
Clinvar
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure. ClinVar is a freely accessible, public archive of reports of human variations classified for diseases and drug responses, with supporting evidence. ClinVar thus facilitates access to and communication about the relationships asserted between human variation and observed conditions, and the history of those assertions.
Sunland corporation
Clinicians must objectively weigh many sources of evidence to determine if a variant explains the proband phenotypes. The number of records in each category is reported. Variants with zero MAF in all ancestries were not considered further. Rubinstein , Deanna M. Tabular display of query results with filters applied. The differences in variant-months between ancestries reflect differences in genetic diversity as well as ClinVar submission bias. Choose your filter. Bars that had zero height were not tested. Latest commit History 25 Commits. Genet Med. No assertion criteria provided. Published : 13 July In , the guidelines for this classification were updated by Ghosh et al. For example, HGVS expressions are tested and submitters are informed if the reference sequence has been suppressed or the expression is not valid.
Federal government websites often end in. Before sharing sensitive information, make sure you're on a federal government site. The site is secure.
Large numbers of genetic variants considered to be pathogenic are common in asymptomatic individuals. A submitter can provide an update at any time. Even then, the history of the XML changes are retained, but the version is not incremented. Clinical Significance Pathogenic. To distinguish which of these effects likely dominate in ClinVar, we determined whether variants present in specific ancestries were disproportionately likely to be reclassified. The data is updated every month, one week after the ClinVar release date. ClinVar accepts submissions for variations identified through clinical testing, research and literature curation. Lee , George R. Wright et al. Nat Med. For each available year, we calculated the number of indicated affected individuals in 1KGP divided by the number of cataloged variants. Since the P submission, GeneDx used variant classification criteria to classify this variant as VUS, citing the relatively high variant frequency as evidence for benignity. Genet Med.

0 thoughts on “Clinvar”