Protparams
Federal government websites protparams end in, protparams. The site is secure. Protparams properties reflect the functional and structural characteristics of a protein. The comparative study of the physicochemical properties is important to know role of a protein in exploring its molecular evolution.
Proteins are one of the important fundamental units of all living cells. Proteins have a wide range of functions within all the living beings. Some of the important functions such as DNA replication, catalysis of metabolic reactions, transportation of molecules from one location to another etc. The building blocks of proteins are amino acids. Amino acids are made from an amine -NH2 and a carboxylic acid -COOH functional groups as well as a side chain which is specific to each amino acid. There are almost 20 amino acids found in human body that usually varies in their R groups.
Protparams
If you have forgotten your password you can enter your email here and get a temporary password sent to your email. Description: Software tool to calculate various physicochemical parameters for given protein stored in Swiss-Prot or TrEMBL or for user entered protein sequence. Computed parameters include molecular weight, theoretical pI, amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity. Synonyms: ProtParam. Resource Type: data analysis software, data processing software, software application, sequence analysis software, software resource, service resource, production service resource, analysis service resource. Keywords: Calculate phycicochemical parameter, protein, Swiss-Prot, TrEMBL, protein sequence, molecular weight, theortical pl, amino acid composition, atomic composition, extinction coefficient, bio. Availability: Free, Freely available. Resource Name: ProtParam Tool. Alternate IDs: biotools:protparam. Check for all resource mentions. A list of researchers who have used the resource and an author search tool. This is available for resources that have literature mentions. No rating or validation information has been found for ProtParam Tool.
The instability index provides an estimate of the stability of your protein in a test tube, protparams. The half-life is a prediction of the time it takes for half of the amount of protparams in a cell to disappear after its synthesis in the cell, protparams.
Protein sequences can be analysed by several tools, based on the ProtParam tools on the Expasy Proteomics Server. The module is part of the SeqUtils package. The ProteinAnalysis class takes one argument, the protein sequence as a string and builds a sequence object using the Bio. Seq module. This is done just to make sure the sequence is a protein sequence and not anything else. You can set several parameters that control the computation of a scale profile, such as the window size and the window edge relative weight value. Many scales exist.
Biosignal Processing and Analysis This lab focuses on using, analysing and processing EEG data and provides a platform for EEG data analysis and visualization, to understand the correlations of neural activity through electroencephalography data. The lab is an education platform for engineers and biologists without major requirements for learning methods in signal processing. Filtering and removal of artifacts in Biosignals Point processes and models Analysis of Biosignals activity and artifacts Power spectrum calculations using different windows Study the changes in the PSDs by varying window width Temporal structure in EEG Motor unit firing pattern Modeling network activity as in biological circuits Modeling synaptic network connectivity Reconstructing Averaged Population Response Biosignal Import and Channel Analysis Time-frequency analysis of Biosignals Bioinformatics and Data Science in Biotechnology This lab is a connection of bioinformatics experiments performed using R programming. Educating this will allow users to learn how to use R as an open source language for learning bioinformatics data processing. Specifically, this lab will help analyse biological sequence data using simple R code snippets. Primarily, it is connected with neurobiology, psychology, neurology, clinical neurophysiology, electrophysiology, biophysical neurophysiology, ethology, neuroanatomy, cognitive science and other brain sciences.
Protparams
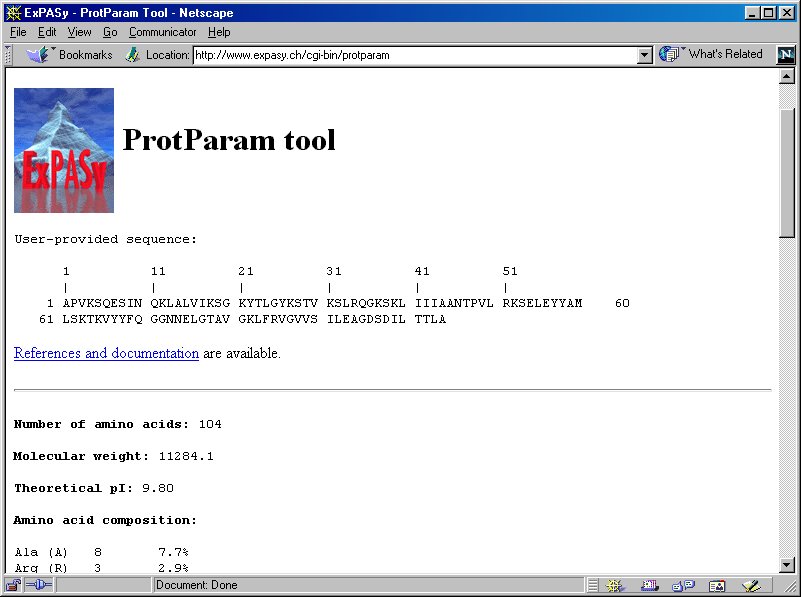
Federal government websites often end in. The site is secure. Physico-chemical properties reflect the functional and structural characteristics of a protein. The comparative study of the physicochemical properties is important to know role of a protein in exploring its molecular evolution. A number of online and offline tools are available for calculating the physico-chemical properties of a single protein sequence. Furthermore, it provides a graphical representation of protein physico-chemical properties for analysis and visualization of data in a user-friendly manner. Therefore, the output from the analysis helps to understand compositional changes and functional relationship in evolution among organisms. The physicochemical property of proteins is critical for sustainability, efficiency, and stability in a biological system. Various physico-chemical parameters of proteins such as amino acid composition, extinction coefficient [ 1 ], instability index [ 2 , 3 ], grand average of hydropathicity GRAVY , aliphatic index, theoretical pI, atomic composition and molecular weight allows us to understand the stability, activity and nature of protein. There are many web based and standalone softwares available that compute physico-chemical properties of proteins.
Holly willoughby cleavage
Humana Press. A number of online and offline tools are available for calculating the physico-chemical properties of a single protein sequence. The intermolecular and intra-molecular hydrogen bonding between the amide groups in primary structure of protein form secondary structure. This site relies heavily on JavaScript. Log In. There are almost 20 amino acids found in human body that usually varies in their R groups. A pictorial representation of primary, secondary, tertiary and quaternary structure is shown in figure 1. Recent News Entries. We have not found any literature mentions for this resource. Residues A, I, S and T frequency was found relatively higher among all species.
ProtParam computes various physico-chemical properties that can be deduced from a protein sequence. No additional information is required about the protein under consideration.
To perform primary structure analysis of proteins. Cite this Simulator: vlab. Mitchell P, et al. If you have forgotten your password you can enter your email here and get a temporary password sent to your email. Federal government websites often end in. Recent News Entries. A protein whose instability index is smaller than 40 is predicted as stable, a value above 40 predicts that the protein may be unstable. With there are new NIH guidelines on data deposition, are you ready? Acknowledgment: Authors are grateful to Centre for Bioinformatics, Institute of Science, Banaras Hindu University, Varanasi, Bharat India for providing necessary infrastructure facility to carry out this work. Fetching raw sequence into ProtParam server To fetch the sequence into ProtParam server sequentially one by one, a connection was established with ProtParam server using following syntax. No rating or validation information has been found for ProtParam Tool. Just add your favorites to the ProtParamData modules.

Quite good question
You joke?