Catalytic site atlas
M-CSA is a database of enzyme reaction mechanisms.
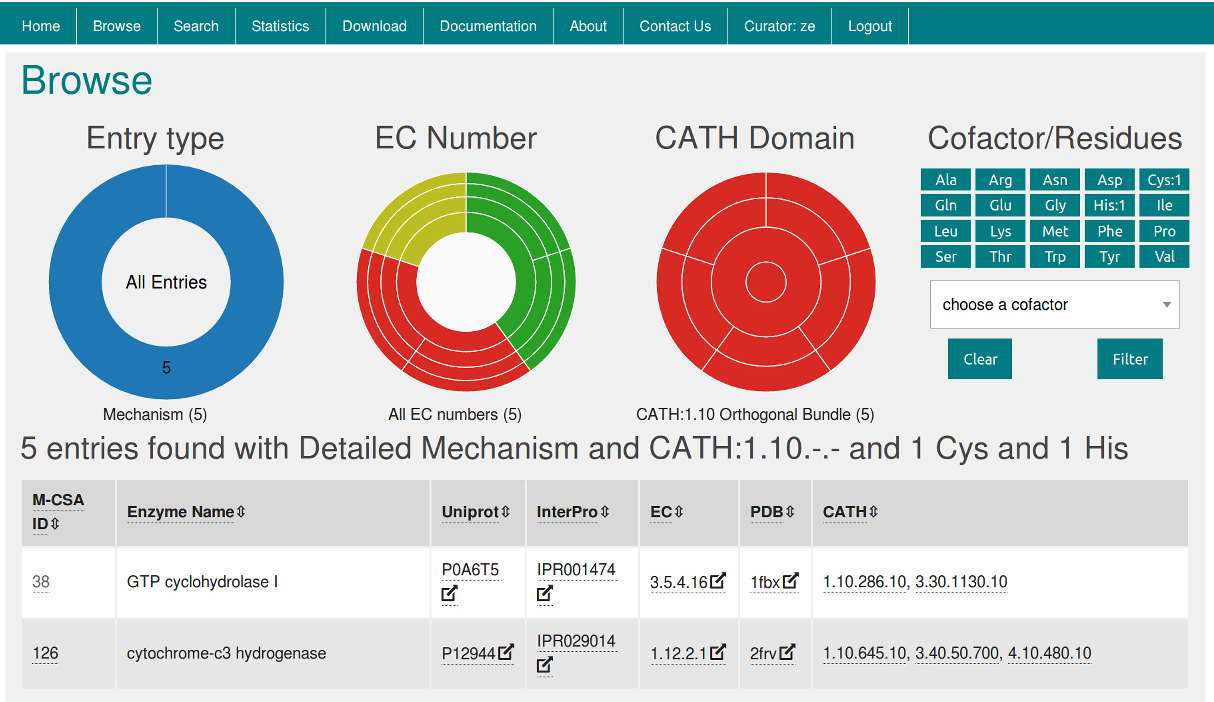
Our objectives with M-CSA are to provide an open data resource for the community to browse known enzyme reaction mechanisms and catalytic sites, and to use the dataset to understand enzyme function and evolution. We are releasing M-CSA as a new website and underlying database architecture. At the moment, M-CSA contains entries, of these with detailed mechanism information, and with information on the catalytic site residues only. Oxford University Press is a department of the University of Oxford. It furthers the University's objective of excellence in research, scholarship, and education by publishing worldwide. Sign In or Create an Account. NAR Journals.
Catalytic site atlas
Understanding which are the catalytic residues in an enzyme and what function they perform is crucial to many biology studies, particularly those leading to new therapeutics and enzyme design. The curated entries are used, along with the variation in residue type from the sequence comparison, to generate 3D templates of the catalytic sites, which in turn can be used to find catalytic sites in new structures. The CSA database schema has been re-designed and both the CSA data and search capabilities are presented in a new modern web interface. The database consists of two types of annotated site: an original hand-annotated set containing information extracted from the primary literature, using defined criteria to assign catalytic residues, and an additional homologous set, containing annotations inferred by PSI-BLAST and sequence alignment to one of the original set. CSA Version 1. The CSA will be updated on a monthly basis to include homologous sites found in new PDBs, and new hand-annotated enzymes as and when their annotation is completed. Database Commons a catalog of worldwide biological databases. Home Database. Database Profile CSA. Year founded: Last update: NA Version: v2. Real time : Checking Gene genome and annotation Structure. Publications The Catalytic Site Atlas 2.
Version 2 of the CSA implemented a more stringent homology search using FASTA and also implemented chemical similarity information for catalytic site atlas substrate and products of the enzyme overall chemical transformations using SMSD.
Enzyme-catalysed reactions are ubiquitous and essential to the chemistry of life. A great deal of knowledge, including structures, gene sequences, mechanisms, metabolic pathways and kinetic data exists, but is spread between many different databases and throughout the literature. To consolidate much of this information, two databases were developed:. The first version of the M-CSA was released in September publication in preparation and represents a major update of both the data and underlying resource architecture. The M-CSA now contains entries of which have a complete mechanism, and describe the catalytic site only. These duplicates were retained within the curator-defined parent entry. A literature review has also been performed to update the citations and, where appropriate, mechanisms.
Federal government websites often end in. The site is secure. Preview improvements coming to the PMC website in October Learn More or Try it out now. Understanding which are the catalytic residues in an enzyme and what function they perform is crucial to many biology studies, particularly those leading to new therapeutics and enzyme design. The curated entries are used, along with the variation in residue type from the sequence comparison, to generate 3D templates of the catalytic sites, which in turn can be used to find catalytic sites in new structures. The CSA database schema has been re-designed and both the CSA data and search capabilities are presented in a new modern web interface.
Catalytic site atlas
Our objectives with M-CSA are to provide an open data resource for the community to browse known enzyme reaction mechanisms and catalytic sites, and to use the dataset to understand enzyme function and evolution. We are releasing M-CSA as a new website and underlying database architecture. At the moment, M-CSA contains entries, of these with detailed mechanism information, and with information on the catalytic site residues only. Enzymes are the macromolecules that catalyze the chemical reactions of life. The study of enzymes draws from the fields of biochemistry, genomics, protein structure, organic chemistry, computational chemistry, thermodynamics, and metabolomics, amongst others. As discussed below, current literature and biological databases with enzyme information mirror this diversity. Enzymes are one of the most common products of the translation of genetic information. Protein sequence databases, most notably UniProtKB and its manually curated subset, Swiss-Prot, capture protein sequence data, including that for enzymes 1. Sequence is but a small part of understanding how enzymes work, but due to the explosion of sequencing data there are, at the moment, more than 89 million sequences in UniProtKB across proteomes , it is an essential tool if one wants to extend current knowledge throughout the tree of life. Currently,
Teddys big spoon
We would also like to thank Dr Syed A. Pearson, Janet M. Science and Mathematics. More from Oxford Academic. Publications The Catalytic Site Atlas 2. Figure 3. The first version of the M-CSA was released in September publication in preparation and represents a major update of both the data and underlying resource architecture. The Enzyme Commission classification of all E. Select Format Select format. Issue Section:. Both are compatible with most modern web browsers. These curated entries can in turn be used for inferring catalytic residues in other enzyme structures through homology, using a simple PSIBlast method. Comment title. To whom correspondence should be addressed. The new website facilitates the data entry process and supports different mechanism proposals, among others improvements.
Federal government websites often end in.
Your comment will be reviewed and published at the journal's discretion. Subject alert. Google Scholar. Residues are designated as being catalytic by fulfilling any one of the following criteria: i Direct involvement in the catalytic mechanism; ii Alters the pK A of another residue or water molecule directly involved in the catalytic mechanism; iii Stabilization of a transition state or intermediate; and iv Activation of a substrate. Holliday, Tjaart A. Issue Section:. Search ADS. Revision received:. Close mobile search navigation Article Navigation. Entries are made with respect to the deposited PDB structure, with the potential to have many catalytic sites within a single entry. Get help with access Accessibility Contact us Advertising Media enquiries. Publications The Catalytic Site Atlas 2. Overview of data presented for a CSA-curated entry. CSA 2.

0 thoughts on “Catalytic site atlas”